(BOSTON) — Synthetic biologists and nanobiologists are re-purposing DNA, the hereditary material present in nearly all the body’s cells, as a smart and stable self-assembling material to build nanofactories, drug-delivering nanostructures and molecular devices that can sense their environment and respond in different ways by, for example, detecting inflammation in the body or toxins in the environment. These nanoscale applications often involve the synthesis of large sequences comprising thousands of the building blocks that DNA is made of, known as the A, T, C and G nucleotide bases, which can be further folded and structured due to the specific base-pairing abilities between As and Ts, and Cs and Gs, respectively.
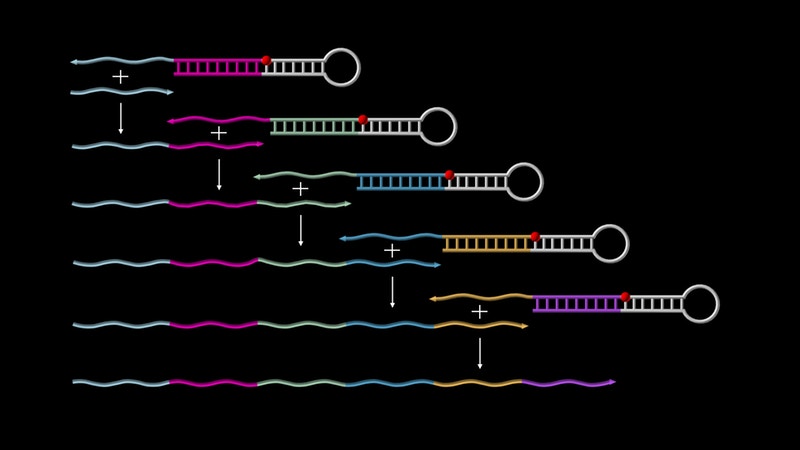
However, so far, researchers do not have tools at their disposal that would allow larger single-stranded sequences to autonomously grow and then join each other end-to-end following a molecular design plan, a capability that could generate structures and devices with diverse capabilities.
ublished today in Nature Chemistry, research by Peng Yin at Harvard’s Wyss Institute for Biologically Inspired Engineering provides a broadly applicable solution to this problem. Yin and his team have developed a method that allows pre-designed sequences of DNA to autonomously grow and concatenate along specific assembly routes, hence providing the basis for a new generation of programmable molecular devices. Putting their new concept of so-called ‘Primer Exchange Reaction’ (PER) cascades to the test, they successfully engineered a first set of devices with diverse functions, such as self-building DNA-origami and DNA nanostructures that sense, amplify, record or logically evaluate environmental signals.
Past methods produced identical copies of a fixed smaller sequence, but they are unable to append different synthesized sequences to each other in defined patterns to generate larger assemblies autonomously without user-mediated intervention. “The autonomous and programmable features that PER cascades offer could engender an entirely new generation of programmable molecular devices and applications and close gaps in design efforts, for which many moving parts already exist,” said Wyss Institute Core Faculty member Peng Yin, Ph.D., who led the study and is also Professor of Systems Biology at Harvard Medical School (HMS). “We provide proof-of-concept data for PER in a diverse range of state-of-the-art synthetic biology applications that clearly highlight the technology’s broad potential.”
The Wyss Institute’s team used the new concept to design a series of such PER DNA transcripts for very diverse applications, including the autonomous synthesis of large DNA nanostructures known as DNA-origamis, and synthetic biology approaches, in which the synthesis of a DNA transcript hinges on a trigger, such as a cancer-associated small micro RNA. Their PER approach can even generate DNA transcripts resulting from a logically evaluated combination of different triggers, similar to RNA Ribocomputing Devices that Yin’s team published earlier this year.
To read more, click: https://wyss.harvard.edu/autonomously-growing-synthetic-dna-strands/
By Benjamin Boettner
